Designing a therapeutic protein that specifically binds its target in drug discovery is a staggering challenge. Traditional workflows are often a painstaking trial-and-error process—iterating through thousands of candidates, each synthesis and validation round taking months if not years. Considering the average human protein is 430 amino acids long, the number of possible designs translates to potential sequences—a practically infinite number, vastly exceeding the number of atoms in the universe (
).
The NVIDIA BioNeMo Blueprint for generative protein binder design is a reference workflow for drug discovery platforms to help them use generative AI and GPU-accelerated microservices to intelligently navigate this immense search space. Instead of brute-force guessing, the system guides to stable, structurally constrained binders, drastically cutting down iterations and time to discovery. This post showcases how researchers at drug discovery companies can rapidly generate novel protein binders, from initial target sequences to validated, stable complexes, all within a streamlined, GPU-accelerated workflow.
Accelerate protein design with NVIDIA NIM and NVIDIA Blueprints
NVIDIA NIM microservices are modular, cloud-native components that accelerate AI model deployment and execution. These microservices enable drug discovery researchers to integrate and scale advanced AI models within their workflows, allowing faster and more efficient processing of complex data.
NVIDIA Blueprints are comprehensive reference workflows that accelerate AI application development and deployment, featuring NVIDIA acceleration libraries, SDKs, and microservices for AI agents, digital twins, and more.
NVIDIA BioNeMo Blueprint for generative protein binder design
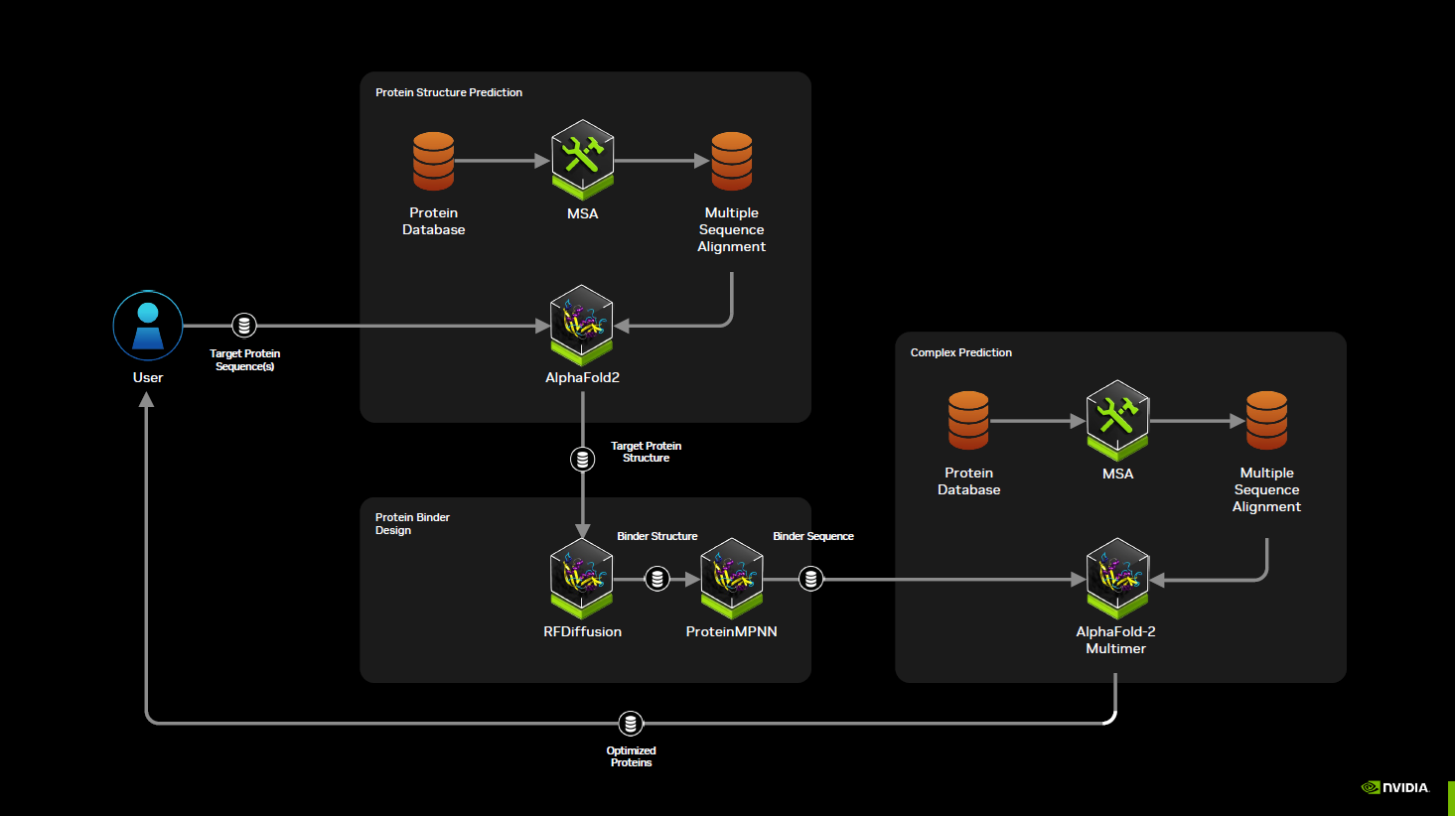
The NVIDIA BioNeMo Blueprint for generative protein binder design provides a comprehensive guide, showing how these microservices can optimize key stages of the protein design workflow.
The process begins with the target protein’s amino acid sequence. This Blueprint seamlessly connects to AlphaFold2 to predict its 3D structure, giving an initial model of what the target looks like.
To aid AlphaFold2’s accuracy, we use an accelerated Multi-Sequence Alignment (MSA) algorithm called MMseqs2 running on NVIDIA GPUs. This ensures a fast, accurate alignment that informs the structure prediction process and enables users to search larger databases that weren’t previously feasible. With MMseqs2 and other upgrades, the AlphaFold2 NIM is now 5x faster and 17x more cost-efficient than the original model.
With the MSA results in hand, AlphaFold2 delivers a 3D model of our target protein. This structure forms the foundation on which we design binders that can latch onto specific regions with high affinity and stability.
Next, the RFdiffusion advanced AI model explores different conformations, guiding us toward optimal binding configurations. Users can fine-tune search parameters to find the best shapes for stable binder-target interactions. With accelerations related to the inference engine, the RFdiffusion NIM is now 1.9x faster than the baseline model.
Once we have a promising conformational landscape, ProteinMPNN takes over. It uses the structural information from RFdiffusion to generate and optimize amino acid sequences that fit these shapes well.
After designing candidate binders, we validate them using AlphaFold2-Multimer. This ensures that the chosen binder and target protein form a stable, well-interacting complex, minimizing the risk of failed experiments downstream.
With these initially validated complexes, researchers can prioritize the most promising candidate protein binder designs, reducing costly and time-consuming lab work. This integrated approach accelerates the design-to-discovery cycle.
Conclusion
Download the NVIDIA BioNeMo Blueprint for generative protein binder design and deploy it anywhere—on-premises, in the cloud, or in hybrid environments. Secure, reliable, and enterprise-supported options can help you scale your research.
With NVIDIA accelerated microservices and generative AI, you can transform protein binder design, boosting efficiency and unlocking new therapeutic possibilities.