Traditional computational drug discovery relies almost exclusively on highly task-specific computational models for hit identification and lead optimization. Adapting these specialized models to new tasks requires substantial time, computational power, and expertise—challenges that grow when researchers simultaneously work across multiple targets or properties.
While specialized models remain widely used, the rise of generalist models has ignited the hope that these versatile frameworks can acquire a useful amount of chemical intuition, meaning that they tackle diverse drug discovery tasks and uncover solutions and patterns that specialized models often overlook.
The recently introduced SAFE-GPT model represented a paradigm shift in AI-driven molecular generation by introducing a chemically intuitive framework aligned with how medicinal chemists approach molecule design. By using the Sequential Attachment-based Fragment Embedding (SAFE) representation (described later in this post), SAFE-GPT addressed critical limitations in earlier molecular generation models to fully capture the flexibility and modularity of molecular structures. This enabled SAFE-GPT to outperform SMILES-based generative models, graph neural networks, and early fragment-based models for a variety of drug discovery-related tasks.
While SAFE-GPT was transformative in its time, it has notable limitations to its efficiency, scalability, and adaptability for diverse drug discovery tasks.
In this post, we compare SAFE-GPT with the recently introduced model, GenMol, presenting the strengths and weaknesses of each and discussing its importance for drug discovery.
SAFE overview
The choice of molecular representation is critically important for the accuracy, efficiency, and versatility of computational models in molecular design and must align with user chemical intuition to become widely adopted.
The SAFE representation, used by both SAFE-GPT and GenMol, reimagines how molecules are described by breaking them into modular, interconnected fragments. Unlike traditional molecular notations like SMILES, which encode molecules as linear strings, SAFE treats molecules as an unordered sequence of fragments. This approach maintains the flexibility and modularity inherent to chemistry while remaining compatible with existing SMILES parsers.
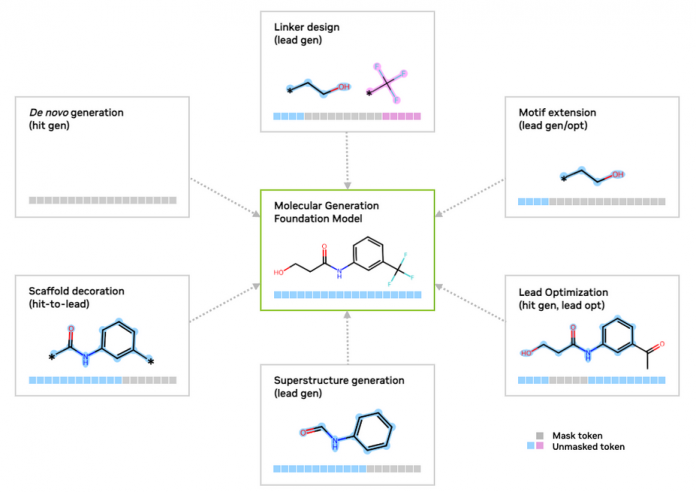
SAFE is particularly well-suited for scaffold decoration, linker design, and motif extension tasks, as it simplifies these problems into sequence completion tasks (Figure 1). By preserving the integrity of molecular scaffolds and accommodating complex structures, SAFE enables intuitive, fragment-based molecular design without the need for intricate graph-based models.
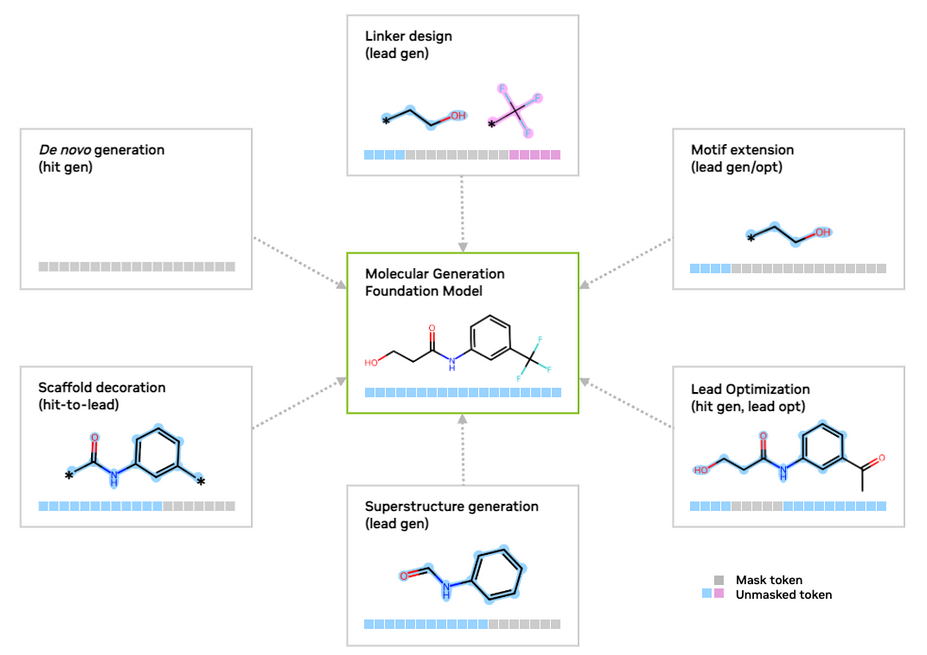
SAFE molecule generations can be used to represent molecules for a variety of tasks relevant to SAFE-GPT and GenMol (Figure 1), including the following:
- Lead optimization: Dynamically replaces molecular fragments with masked tokens to refine and optimize designs iteratively.
- De novo generation: Creates entirely new molecular structures from scratch, enabling the discovery of novel compounds with desired properties.
- Linker design: Optimizes molecular linkers that connect functional fragments, enhancing structural stability and activity.
- Motif extension: Extends key molecular motifs to explore additional functional groups or enhance target binding interactions.
- Superstructure generation: Constructs complex, multi-fragment molecular architectures for advanced drug discovery and material design.
- Scaffold decoration: Modifies core molecular scaffolds by adding diverse substituents to explore structure-activity relationships.
Example GenMol inference code
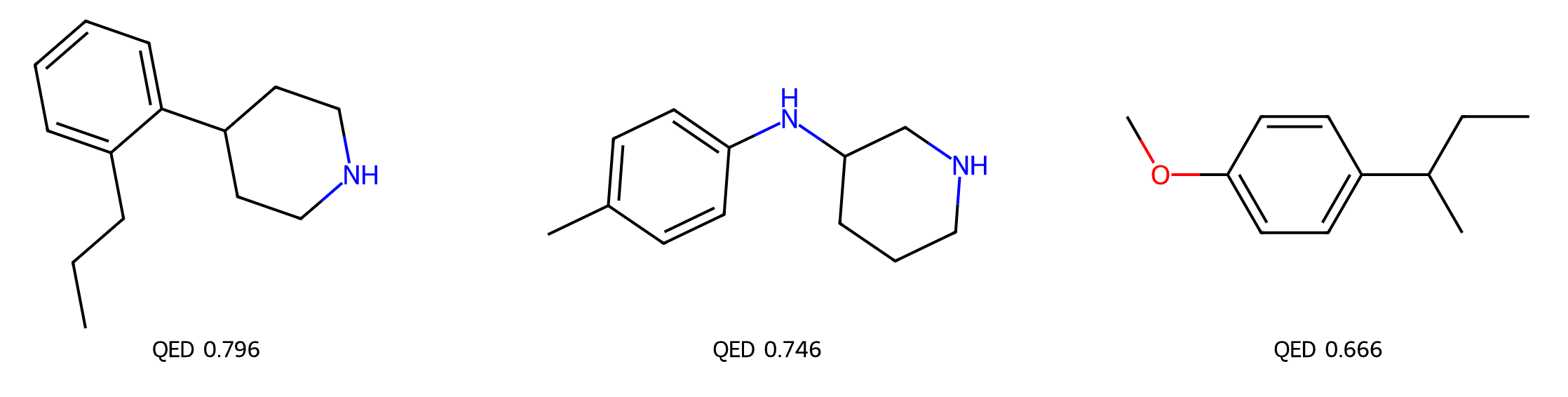
The GenMol NIM microservice and its companion notebooks simplify inference requests by enabling you to input varying SAFE or SMILES and mask strings, with de novo generation requiring only a pure mask and the desired molecule count:
generator = GenMol_Generator(invoke_url='http://127.0.0.1:8000/generate)
# provide a SMILES or a SAFE sequence string
molecules = generator.inference(smiles='[*{15-25}]',num_molecules=20)
Figure 2 shows the example output.
For linker design or motif extension, you can provide a mask for a set of fragments by appending or inserting the mask into those fragments:
# append a mask
input_text = 'c14ncnc2[nH]ccc12.C136CN5C1.S5(=O)(=O)CC.C6C#N.[*{15-35}]'
# or insert the mask
input text = 'c14ncnc2[nH]ccc12.C136CN5C1.[*{5-15}].S5(=O)(=O)CC.C6C#N'
# generate molecules
# provide a SMILES or a SAFE sequence string
molecules = genmol.inference(smiles=input_text,temperature=1.5,
noise=1.0,num_molecules=1000)
Figure 3 shows the example output.
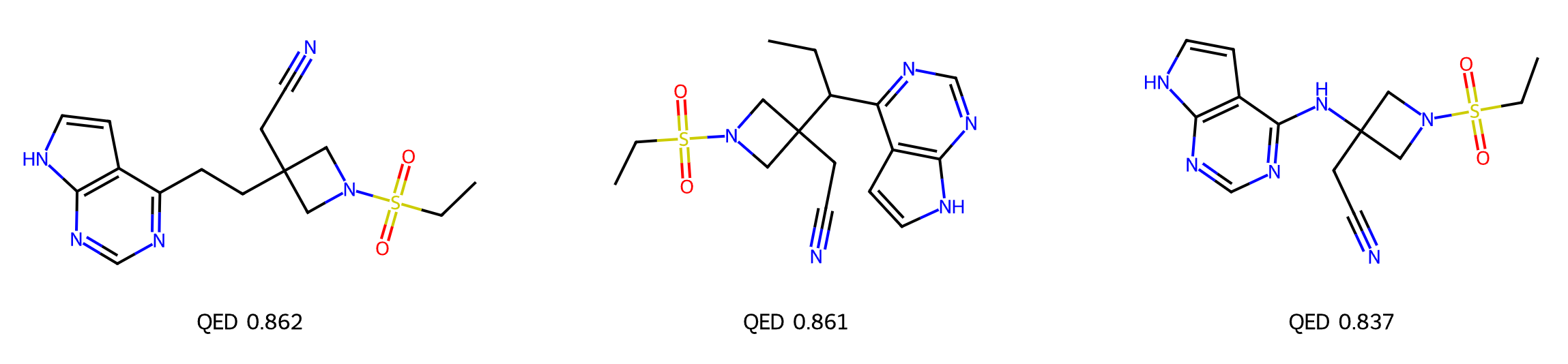
By introducing a fragment library and a QED-scoring oracle, you can iteratively use GenMol inference for advanced guided optimization, enabling hit generation and lead optimization based on input mask structures.
library = Library(max_fragments = 1000)
oracle = Oracle(score = Oracle.RDKitScore('QED'))
optimizer = MolecularOptimizer(library = library, oracle = oracle,
generator = generator)
data = []
def eval():
best = library.molecules.score[0]
mean = library.molecules.head(50)['score'].mean()
std = library.molecules.head(50)['score'].std()
print("BEST", best, "MEAN", mean, "STD", std)
data.append([best, mean, std])
eval()
for i in range(100):
optimizer.run(iterations = 10, num_mutate = 50)
eval()
By iteratively optimizing and updating the fragment library with high-scoring candidates—exemplified here with QED but applicable to any property or combination of properties—you can quickly guide the generative process.
Comparing SAFE-GPT and GenMol for drug discovery tasks
GenMol and SAFE-GPT represent two distinct approaches to AI-driven molecular generation, each with unique strengths and limitations (Table 1).
| Feature | GenMol | SAFE-GPT |
| Decoding | Parallel (non-autoregressive) | Sequential (autoregressive) |
| Task versatility | Broad | Requires task-specific adaptation |
| Efficiency | Scalable and efficient | Computationally intensive |
| Diversity-quality trade-off | High balance | Moderate |
SAFE-GPT, built on an autoregressive transformer architecture, is a powerful tool for fragment-constrained tasks, such as scaffold decoration and linker design. Its use of sequential decoding ensures precision and chemical validity in these specific scenarios. However, its sequential nature and task-specific design can be computationally intensive and less adaptable to new tasks without retraining.
GenMol, with its discrete diffusion-based architecture and parallel decoding, addresses many limitations by improving computational efficiency and task versatility. It extends the scope of molecular generation to include broader challenges such as goal-directed lead optimization, where it outperforms even widely used models such as f-RAG and REINVENT. Its dynamic fragment remasking strategy enables robust exploration of chemical space, making it suitable for more complex, multi-objective drug discovery workflows.
Beyond goal-directed lead optimization, each model’s decoding strategy influences its performance in fragment-based tasks, as we will see next.
Molecular generation and exploration of chemical space
SAFE-GPT uses a GPT architecture with sequential, autoregressive decoding, generating molecules fragment-by-fragment. SAFE-GPT, combined with the fragment order-insensitive nature of the SAFE representation, can be applied to de novo and fragment-constrained generation of molecules.
GenMol, built on a BERT architecture, employs parallel, non-autoregressive decoding with bidirectional attention, processing molecular fragments simultaneously. This enables GenMol to consider the molecular context that does not rely on the arbitrary ordering of tokens and fragments and notably outperforms SAFE-GPT in fragment-constrained molecule generation tasks as measured by their quality score (Table 2).
| Task | SAFE-GPT | GenMol |
| Motif extension | 18.6% +- 2.1 | 27.5% +- 0.8 |
| Scaffold decoration | 10.0% +- 1.4 | 29.6% +- 0.8 |
| Superstructure generation | 14.3% +- 3.7 | 33.3% +- 1.6 |
Furthermore, discrete diffusion enables GenMol to explore chemical space using a fragment-remasking strategy, which dynamically replaces fragments with masked tokens, enhancing its ability to discover novel and optimized molecules through iterative refinement. This enables GenMol to be applicable to hit generation and lead optimization tasks without any task-specific fine-tuning.
Computational efficiency
SAFE-GPT’s sequential generation and reliance on reinforcement learning objectives make it computationally intensive, particularly for large-scale or high-throughput scenarios.
GenMol’s discrete diffusion framework improves generation efficiency, offering up to 35% faster sampling and lower computational overhead, making it more scalable for industrial-scale drug discovery.
Conclusion
The importance of these molecular generation models goes beyond just how molecular generation is done. It also explains why it needs to be reimagined.
In an industry where time-to-market can mean the difference between life and death for patients, more broadly useful models can equip researchers with a versatile, efficient, and precise tool to streamline discovery, optimize outcomes, and expand the horizons of what is chemically possible. They represent a critical leap forward from labor-intensive processes to AI-driven innovation that is as adaptive as the challenges it seeks to solve.
Both models provide valuable tools depending on the specific needs of a research project. SAFE-GPT is an excellent choice for projects focused on motif extension and scaffold generation with strict fragment constraints, while GenMol is better suited for researchers requiring a more flexible, unified framework for diverse drug discovery applications.
Test GenMol as an NVIDIA NIM now or explore code examples on GitHub to learn more about using GenMol for goal-directed hit optimization, lead optimization, and more. Explore these approaches further to determine the best fit for your research needs and accelerate your work in drug discovery.